Why does it bother?
It seems like a peculiar case of genomic overkill: a single-celled bacterium has been found that keeps tens of thousands of copies of its genome. The finding sets a record for most genomes per cell, but also poses an obvious question: what could be the advantage of stashing away as much as 200,000 copies of your genome?
The number of genome copies in each cell varies by species. Many bacteria have only one copy; most cells in the human body contain two. Plants are notorious for being genomically promiscuous, picking up extra genomes then losing them again in a cycle that can occur repeatedly in a plant’s evolutionary history. “You may think that as a blueprint of life, the genome might be very stable in size,” says Sally Otto, an evolutionary biologist at the University of British Columbia in Vancouver, Canada. “So it is really quite remarkable that organisms vary so widely in the number of genome copies they have.”
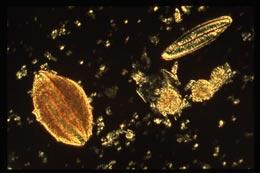 More genomes than any predator would want to swallowEsther Angert
More genomes than any predator would want to swallowEsther AngertUntil now, the record for the ‘most genomes’ in a bacterium was held by Buchnera aphidicola, which is found in aphids and carts around an average of 120 genomes. But Esther Angert of Cornell University in Ithaca, New York, and her colleagues now report in the Proceedings of the National Academy of Sciences 1 that Epulopiscium, a gargantuan bacterium that lives in the intestines of unicornfish, contains up to 200,000 copies of its genome.
Living large
Angert first heard of Epulopiscium as a graduate student while doing a rotation in Norman Pace’s lab at Indiana University in Bloomington. She asked whether she could stay in the lab to study lichen. Pace pulled a sample of Epulopiscium out of the refrigerator and suggested: ‘how about working on these?’ Angert took one look at them under the microscope and was hooked: “They are beautiful organisms,” she says. “They are elegant.”
Soon, Angert found herself harvesting unicornfish guts to analyse DNA sequences from Epulopiscium . At the time, the gigantic Epulopiscium with its hairy flagellar coat and what seemed to be internal structures bound to the membrane, was classified as a ‘protist’: a junk-pile of a category populated with eukaryotes that don't fit into any better-defined clades. Angert’s analysis grouped Epulopiscium with bacteria. The internal structures, it turned out, were offspring starting to grow while still inside their parent.
Epulopiscium can be up to 600 micrometres long — much larger than most other bacteria. Cigar-shaped and tan in colour, the bacteria look like a tiny splinter to the naked eye. Escherichia coli, the familiar bacteria that colonizes human intestines, is about 1 micrometre long. “You could easily fit a million E. coli cells in Epulopiscium ,” Angert says.
Gigantism is usually a disadvantage in bacteria because they do not have the sophisticated nutrient uptake systems characteristic of eukaryotic cells. Instead, bacteria harvest nutrients via simple diffusion across the cell membrane. Because a large bacterium will have a lower ratio of surface area to cell volume, the rate at which it takes on nutrients might not be high enough to keep it alive. Large bacterial cells would simply starve.
The intestinal Garden of Eden
But being big has its advantages, notably that it deters predation. Epulopiscium is too big for other microbes to prey on; Angert says that she has seen only one microbe pull off that feat: the ciliate Balantidium jocularum . “They really have to stretch to get some of these guys into their ‘mouths’, but they manage to do it,” she says.
Angert suspects that Epulopiscium ’s extravagant collection of genomes may be a way for it to reap the benefits of size without the drawbacks of starvation. She hypothesizes that the genomes are arrayed just beneath Epulopiscium ’s cell membrane. This arrangement means that the cell could respond to nutrients and other environmental molecules without waiting for them to diffuse throughout the cell. “If you waited for an environmental signal to get to you, relying solely on diffusion, it would take forever,” says Anger. “It would be really unreliable. This opens up that door of allowing the cell to get big and not allowing diffusion to limit its volume.”
Having so many genomes also ramps up the rate at which Epulopiscium can produce RNA and protein from its genes, a hypothesis favoured by microbiologist Hank Siefert of Northwestern University in Chicago, Illinois, who says that such a system could help keep the gargantuan bacterium running smoothly.
What about other behemoth bacteria? Epulopiscium is not the largest bacterium known: Thiomargarita namibiensis , so named for its round, pearly sulphur granules rather than because it makes a good cocktail, grows up to 800 micrometres long. So far no one has tallied the number of genomes in a T. namibiensis cell, but the bacterium contains a large central structure called a vacuole that is filled with fluid. The vacuole takes up 98% of the cell volume in T. namibiensis, shoving the metabolically active parts of the cell up against the cell membrane. In Epulopiscium, on the other hand, the central contents of the cell are also active, and would require a supply of nutrients and proteins.
It takes a significant investment of energy to maintain all those genomes, but the lush environment of the unicornfish gut may make that trade-off easier to bear, says Seifert.



No comments:
Post a Comment